CLASSIX is a fast, memory-efficient, and explainable clustering algorithm. Here are a few highlights:
- Ability to cluster low and high-dimensional data of arbitrary shape efficiently
- Ability to detect and deal with outliers in the data
- Ability to provide textual and visual explanations for the clusters
- Full reproducibility of all tests in the accompanying paper
- Support of Cython compilation
CLASSIX is a contrived acronym of CLustering by Aggregation with Sorting-based Indexing and the letter X for explainability.
CLASSIX can be installed via PIP (recommended) or Conda:
| PyPI | conda-forge |
|---|---|
NumPy<=1.26.4: pip install classixclustering NumPy>2: pip install classixclustering --no-cache-dir |
conda install -c conda-forge classixclustering |
We recommend the installation with pip install classixclustering --no-cache-dir .
| Language | Dependencies |
|---|---|
 |
    |
Here is an example of clustering one of the demo dataset provided with CLASSIX:
import classix
data, labels = classix.loadData('Covid3MC')
# or one can use Gaussian blobs for a quick try:
# from sklearn.datasets import make_blobs
# data, labels make_blobs(n_samples=1000, centers=3, n_features=2, random_state=0)
# Call CLASSIX
clx = classix.CLASSIX(radius=0.2, minPts=500, verbose=0)
clx.fit(data)
print(clx.labels_) # clustering labels You can also cluster out-of-sample data using predict() after the model is fitted, e.g., clx.predict(data.iloc[:1000])
CLASSIX is an explainable clustering method. To get an overview of the computed clusters, type:
clx.explain()Output:
CLASSIX clustered 5726839 data points with 3 features.
The radius parameter was set to 0.20 and minPts was set to 500.
As the provided data was auto scaled by a factor of 1/0.33,
points within a radius R=0.20*0.33=0.07 were grouped together.
In total, 80596471 distances were computed (14.1 per data point).
This resulted in 301 groups, each with a unique group center.
These 301 groups were subsequently merged into 25 clusters.
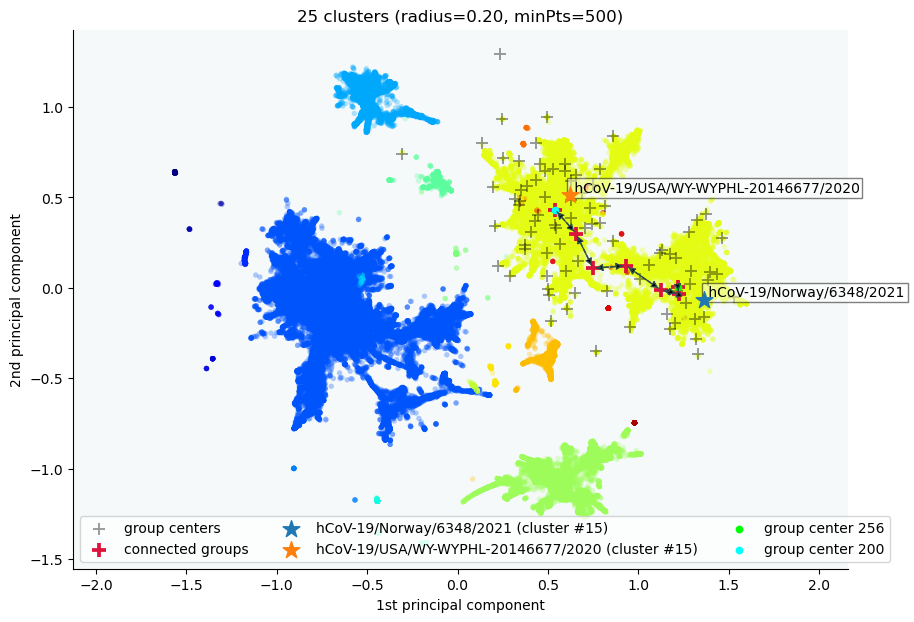
We can ask CLASSIX why two data points ended up in the same cluster, or not:
clx.explain('hCoV-19/Norway/6348/2021','hCoV-19/USA/WY-WYPHL-20146677/2020', plot=True) # one can also provide native integer indicesOutput:
Data point hCoV-19/Norway/6348/2021 is in group 256.
Data point hCoV-19/USA/WY-WYPHL-20146677/2020 is in group 200.
Both groups were merged into cluster #15.
The two groups are connected via groups
256 <-> 257 <-> 248 <-> 242 <-> 225 <-> 217 <-> 200.
Here is a list of connected data points with
their global data indices and group numbers:
Index Distance Group Label
600 -- 256 hCoV-19/Norway/6348/2021
186838 0.17 256 hCoV-19/Ireland/LD-NVRL-89IRL62956/2021
5261957 0.28 257 hCoV-19/Northern_Ireland/NIRE-003afb/2021
5086569 0.20 248 hCoV-19/Ukraine/Kyiv-PHC-0002/2021
4600143 0.23 242 hCoV-19/Belgium/rega-2620/2021
1599046 0.20 225 hCoV-19/Germany/NW-RKI-I-088626/2021
273188 0.21 217 hCoV-19/USA/CA-ALSR-8586/2020
167935 0.24 200 hCoV-19/Spain/CT-HUB00588/2021
100 0.13 200 hCoV-19/USA/WY-WYPHL-20146677/2020
The distance between consecutive data points is at most R=3.68.
Here, R=0.2*12.27*1.5, where 0.2 is the chosen radius parameter,
dataScale_=12.27 is a data scaling factor determined by CLASSIX,
and mergeScale_=1.5 (the default value).
CLASSIX clustering consists of two phases, namely a greedy aggregation phase of the data into groups of nearby data points, followed by a merging phase of groups into clusters. The radius parameter controls the size of the groups and minPts controls the minimal cluster size. CLASSIX explains that there is a path from data point 'hCoV-19/Costa_Rica/HNN-0400/2021' to data point 'hCoV-19/Denmark/DCGC-269073/2021' via the centers of the computed groups ( 191, 210, etc).
CLASSIX supports dataframes and using dataframe indices to refer to data points:
from sklearn import datasets
from classix import CLASSIX
import pandas as pd
X, _ = datasets.make_blobs(n_samples=5, centers=2, n_features=2, cluster_std=1.5, random_state=1)
X = pd.DataFrame(X, index=['Anna', 'Bert', 'Carl', 'Tom', 'Bob'])
# 0 | 1
# Anna | -7.804551 | -7.043560
# Bert | -9.519154 | -4.327404
# Carl | -0.361448 | 0.954182
# Tom | 0.957658 | 3.264680
# Bob | -2.451818 | 2.797037
clx = CLASSIX(radius=0.6)
clx.fit_transform(X)
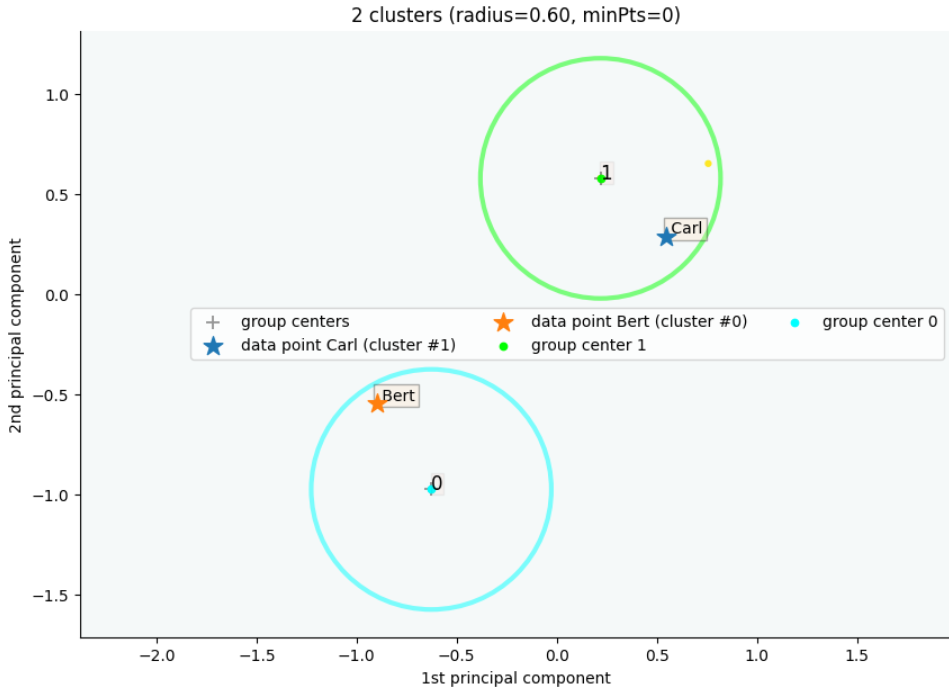
clx.explain(index1='Carl', index2='Bert', plot=True, showallgroups=True, sp_fontsize=12)Output:
The data point Carl is in group 1, which has been merged into cluster 1.
The data point Bert is in group 0, which has been merged into cluster 0.
There is no path of overlapping groups between these clusters.
We recommend first running CLASSIX with a relatively large radius parameter, such as radius=1, and minPts=1. It also helps to use verbose=1 to get more detailed feedback from the method. Typically, the larger the radius parameter, the faster the method performs and the smaller the number of computed clusters. If the number of clusters is too small, successively reduce the radius parameter until a "good" (depending on context) number of meaningful clusters is obtained.
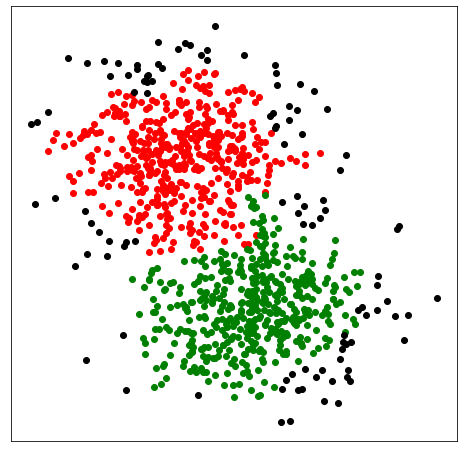
One can access the size of the clusters via clx.clusterSizes_. If there are unwanted "noise" clusters containing just a small number of data points, increase the minPts parameter to remove them. If, for example, minPts=14, all clusters with fewer than 14 data points will be reassigned to larger clusters. Here is an example that demonstrates the effect of minPts:
# generate synthetic data
X, y = datasets.make_blobs(n_samples=1000, centers=2, n_features=2, random_state=0)
# run CLASSIX
clx = CLASSIX(sorting='pca', radius=0.15, group_merging='density', verbose=1, minPts=14, post_alloc=False)
clx.fit(X)
# (omit plot code)Output:
CLASSIX(sorting='pca', radius=0.15, minPts=14, group_merging='density')
The 1000 data points were aggregated into 212 groups.
In total 5675 comparisons were required (5.67 comparisons per data point).
The 212 groups were merged into 41 clusters with the following sizes:
* cluster 0 : 454
* cluster 1 : 440
* cluster 2 : 13
* cluster 3 : 10
* cluster 4 : 7
--- lines omitted ---
* cluster 38 : 1
* cluster 39 : 1
* cluster 40 : 1
As MinPts is 14, the number of clusters has been further reduced to 3.
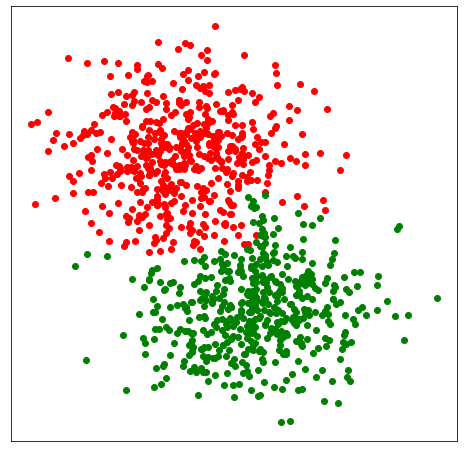
Note that there are many clusters with fewer than 14 data points. Because minPts=14 and post_alloc=False all of these tiny clusters are labelled as noise with the label -1. We can also reallocate noisy clusters to their respective nearby clusters by setting post_alloc=True (which is the default value). In this case we get the following clustering:
When there are many data points, the visualisations produced by the .explain method might be difficult to interpret. There are several options that help producing better plots, e.g. when the boxes of starting points are too large so that they hide the data points. In this case, we may set sp_alpha smaller to get more transparency for the box of starting points or set sp_pad smaller to get the box smaller, or we can change the color of that by specifying sp_fcolor to a more shallow color. For more detail, we refer users to the documentation. Also, you can set cmap (e.g., 'bmh'), cmin and cmax to customize a color map of the clusters.
CLASSIX used Cython to speed up some parts of its computation. If you get a Cython warning, it means that your current Python installation does not support Cython, or there is no C compiler present on the system. Getting a C compiler varies according to the system used, e.g., GNU C Compiler (gcc) for Linux, Microsoft Visual C/C++ (MSVC) for Windows. CLASSIX will run just fine without Cython, but it might be slower than usual (sometimes up to a factor of 40). To double check whether you are using Cython or not, please use:
import classix
classix.cython_is_available(verbose=1)If needed, Cython can be disabled via
classix.__enable_cython__ = FalsePlease refer to the CLASSIX paper referenced below. In short, CLASSIX sorts the data points along their first principal axis and then aggregates them into groups of radius radius. This group merging is sped up by exploiting high-level BLAS best practices and using an early stopping criterion to terminate the group search. The computed groups are then merged into clusters. In a final stage, the groups of any clusters with fewer than minPts points will be reallocated to larger clusters.
Here is short video abstract on CLASSIX:
A detailed documentation, including tutorials, is available at 
demos subfolder.
All experiments in the CLASSIX paper referenced below are reproducible by running the code in the folder of "exps".
Before running, ensure the dependencies scikit-learn and hdbscan are installed and the Quickshift++ code (Quickshift++: Provably Good Initializations for Sample-Based Mean Shift) is compiled. After configuring all of these, run the following commands:
cd exp
python3 run exp_main.py
All results will be written to the folder "exps/results". Please let us know if you have any questions.
Any form of contribution is welcome. We particularly welcome the contribution of new demos in the form of Jupyter Notebooks. Feel free to post issues and pull requests if you want to assist in documentation or code. To contribute, please fork the project and pull a request for your changes. We will strive to work through any issues and requests and get your code merged into the main branch. Contributors will be acknowledged in the release notes.
@article{CG24,
title = {Fast and explainable clustering based on sorting},
author = {Xinye Chen and Stefan Güttel},
journal = {Pattern Recognition},
volume = {150},
pages = {110298},
year = {2024},
doi = {https://doi.org/10.1016/j.patcog.2024.110298}
}This project is licensed under the terms of the MIT license.